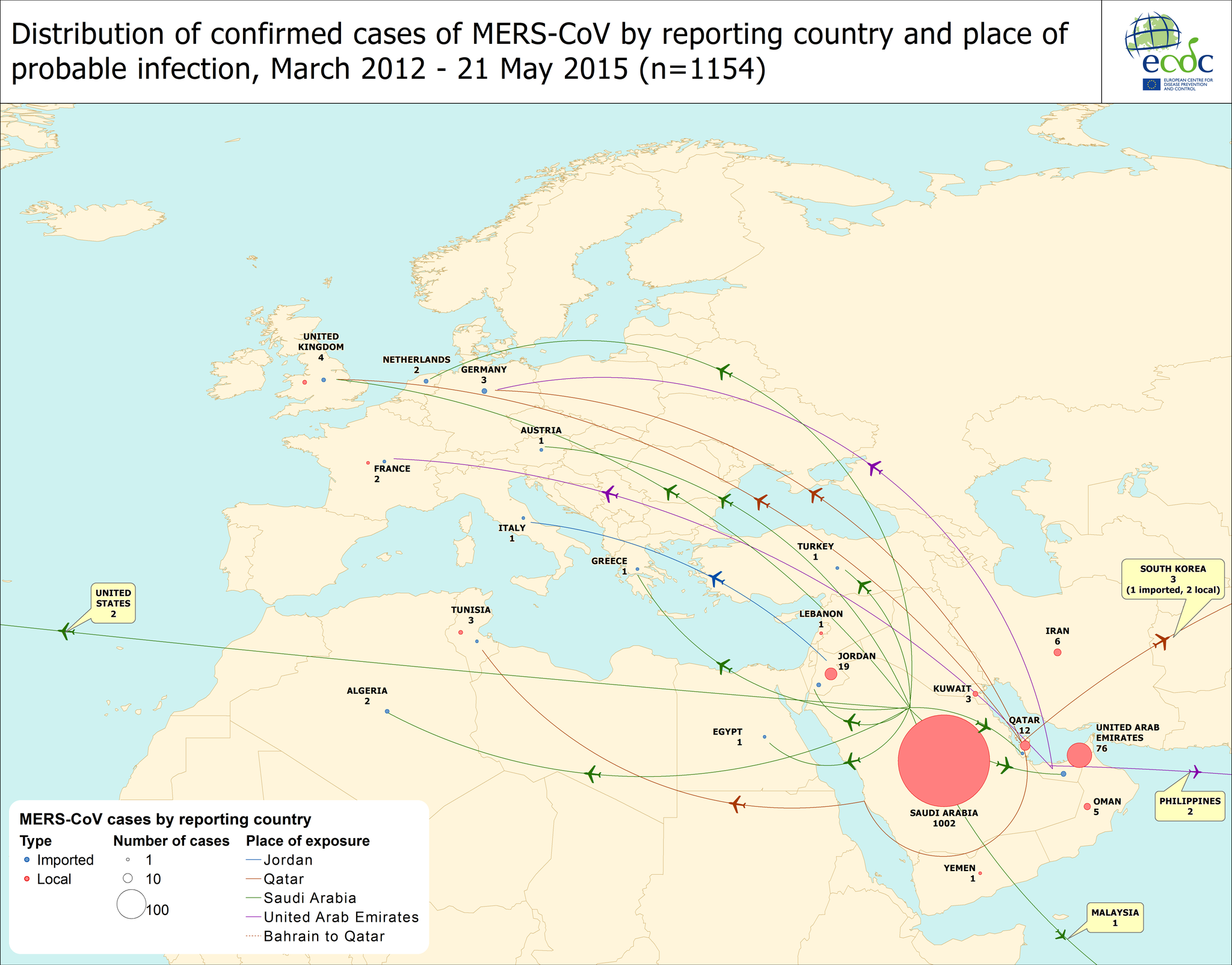
The authors remind us in the background that the routes of direct or indirect zoonotic transmission are still unknown but that a "large proportion of MERS cases" are suspected to have resulted from zoonotic transmission.
105 dromedary camels (DCs) either from a market sale or directly from Qatar or the Kingdom of Saudi Arabia (KSA) were sampled in February (n=53) and March (n=52), 2014. Samples included nasal, oral, rectal and bronchial swabs and lymph nodes from animals grouped into age 3 groups: 0 to 6 months (n=41), 7 to 12 months (n=35) or greater than 12 months (n=29) of age. Testing for virus was by Corman et al's UpE and N gene real-time RT-PCRs.[2] Testing for antibodies was via the detection of a reaction to the MERS-CoV, severe acute respiratory syndrome (SARS)-CoV and human CoV (HCoV)-OC43 spike domain S1 antigen using the protein-microarray method described previously by this group.[4]
Findings...
- 59% of DCs had at least one MERS-CoV RNA positive sample but no significant difference in viral load was apparent between sample types or ages
- 61/101 (60.3%) of DC's nasal samples had RNA detected
- 23/102 (22.5%) of DC's saliva samples had RNA detected
- 15/103 (14.6%) of DC's rectal samples had RNA detected
- 7/101 (6.9%) of DC's bronchial samples had RNA detected
- 5/53 (9.4%) of DC's lymph nodes had RNA detected
- 5 different MERS-CoV variants (subtly different versions of MERS-CoV) were circulating in Qatar among the sampled animals at this time according to RT-PCR/sequencing method that targets a fragment of the S2 domain of the MERS-CoV Spike gene.[3]
- 100/103 (97%) animals were reactive for IgG, and most of 53 animals tested, had antibodies capable of specifically neutralizing cellular infection by MERS-CoV as determined by a 90% plaque reduction neutralization test (PRNT90; [5])
- Antibody levels and viral load did not correlate suggesting - based on this subset of the immune response - that reinfection may be possible since protection may be limited, as it is among humans with the 4 known HCoVs. The authors note that this may prove a challenge for any future DC vaccine which would need to produce a protective effect to meets its need
- No age-specific differences were found in MERS-CoV RNA shedding - usually younger DCs are distinctly more likely to be shedding viral RNA than older DCs
Discussion...
The authors noted here that discrepancies do exist between their study and those of some others - specifically, that others have not found viral RNA in faeces - but those studies also tested fewer animals. It is important, when percentages are not high, to test enough animals to see the full extent of MERS-CoV shedding and potential transmission routes.
DCs from different regions within Qatar and outside Qatar, may be shedding MERS-CoV while in DC markets and holding pens, sometimes for weeks, awaiting slaughter.
Camel markets are thus a likely high risk area for acquiring a MERS-CoV infection - and multiple variants can be circulating here.
In previous Qatari investigations, human cases have been linked with visits to the areas studied here and have also included DC slaughterer cases, supporting the notion that humans with DC exposures (presumably when they are infected with MERS-CoV) are at risk of becoming infected themselves.
Yet this study did not manage to capture the process of transmission in action. It is that process that holds such importance for this chapter on MERS-CoV and especially for those who disbelieve the role of DCs in human MERS cases.
In the next post, we will re-visit a study that did seem to capture DC>human infection.
- High proportion of MERS-CoV shedding dromedaries at slaughterhouse
with a potential epidemiological link to human cases, Qatar 2014.
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4505336/ - http://www.ncbi.nlm.nih.gov/pubmed/23041020
- http://www.ncbi.nlm.nih.gov/pubmed/25728084
- http://virologydownunder.blogspot.com.au/2015/10/if-you-are-often-in-contact-with-camels.html
- http://www.thelancet.com/journals/laninf/article/PIIS1473-3099(13)70164-6/abstract