Edited for clarity 25MAY2015
Middle East respiratory syndrome coronavirus (MERS-CoV) managed to get out for some sightseeing - travelling to South Korea this week - and Ebola virus|Makona was given a helping hand to spread to new people in Guinea and Sierra Leone with a small splurge of new confirmed cases.
MERS has now trickled into 24 countries world wide as shown in the European Centre for Disease Prevention and Control's (ECDC) epic 'travel-by-plane' map.
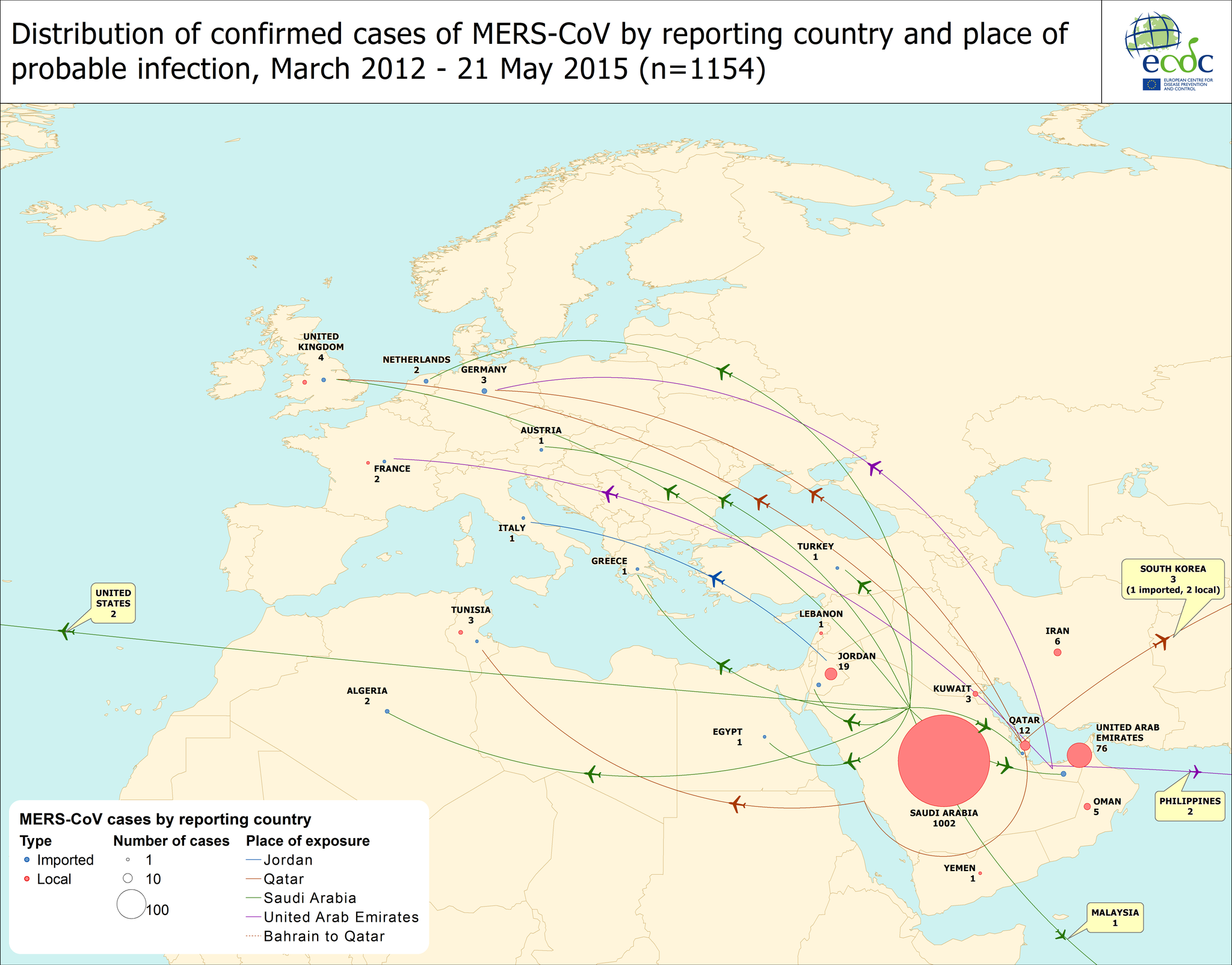 |
| The original of this is created by the ECDC and is presented here. Click on image to enlarge. |
Meanwhile, a crude extrapolation from current Ebola virus disease (EVD) case numbers saw the predicted date when we might reach zero cases, move further into June.
This could pull back again or it could move further away if the EVD clusters and sporadic cases continue to spread. We can't model that because it's entirely down to unpredictable human variables. We can list what those are, we can better prepare for them, we can educate about them and how to prevent them and we can acknowledge that they are real, but we cannot know when and in what mix they will come into play.
This could pull back again or it could move further away if the EVD clusters and sporadic cases continue to spread. We can't model that because it's entirely down to unpredictable human variables. We can list what those are, we can better prepare for them, we can educate about them and how to prevent them and we can acknowledge that they are real, but we cannot know when and in what mix they will come into play.
 |
| Extrapolation of the public data for confirmed Ebola virus disease cases from WHO. The most recent week is boxed in red and bucked the trend of declining cases. To see how I made this please visit here. Click on image to enlarge. |
The newest EVD cases remain mostly clustered around the Forecariah prefecture of western Guinea, on the north west border with Sierra Leone but also 5 new cases appeared in the north west of Guinea in Boke prefecture, which borders Guinea-Bissau.
| From the World Health Organization's Ebola virus disease Situation Report, 20MAY2015. Click on image to enlarge. |
Since the last EVD SitRep, two days of reporting have seen fewer cases than in the same two days of the week before.
So there's that.
Quickly reporting what is actually happening is invaluable for all sorts of reasons. Modelling and prediction allow us to get ahead of the virus. But having the data, and having them available publicly remains a challenge for every country and for every outbreak.
So there's that.
Quickly reporting what is actually happening is invaluable for all sorts of reasons. Modelling and prediction allow us to get ahead of the virus. But having the data, and having them available publicly remains a challenge for every country and for every outbreak.
Public health data are about the public's health. If it has been considered worth collecting and collating, why not communicate it too?



